About
I am an Instructor in Medicine in the Division of Preventive Medicine at Brigham and Women’s Hospital
(BWH) and Harvard Medical School (HMS), and I am a member of
Melina Claussnitzer,
Ph.D lab at the Broad Institute of MIT and Harvard and Massachusetts General Hospital.
My effort at the Broad Institute is focused on using advanced computational methods for the conversion
of disease-associated genetic variants to function (V2F). These genetic V2F studies are focused on
cardio-metabolic diseases, where we utilize advanced computational methods to investigate a variety of
high-dimensional cellular and molecular phenotypic data, including chromatin accessibility, gene
expression and high content imaging-based profiling data at the single cell level.
In collaboration with
Kathryn Hall, Ph.D.,M.P.H., at BWH and HMS, we are working on pharmacogenomics and precision medicine.
In this collaboration, we utilize advanced computational methods to investigate precision efficacy of
drugs and dietary supplements in CVD and cancer.
In collaboration with Samia Mora, M.D. and
Olga Demler, Ph.D.
my efforts at BWH and HMS are in computational CVD, with a focus on using machine learning methods for
predicting medical events. In addition, we identify and determine levels of disease risk biomarkers
using nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS).
Web-servers
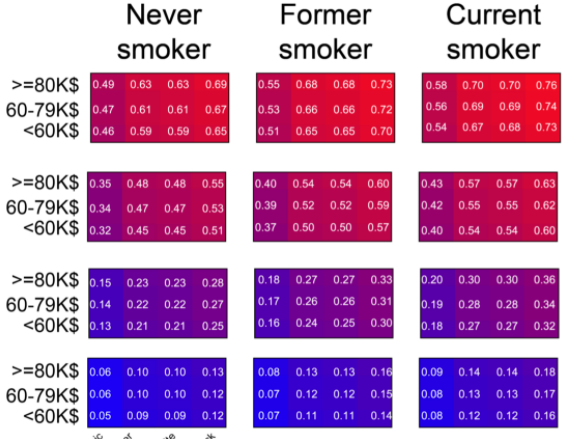
SARS2
Simplified scores to estimate risk of hospitalization and death among patients with COVID-19 based on Sex, Age, Race, Socioeconomic, Smoking status (SARS2).
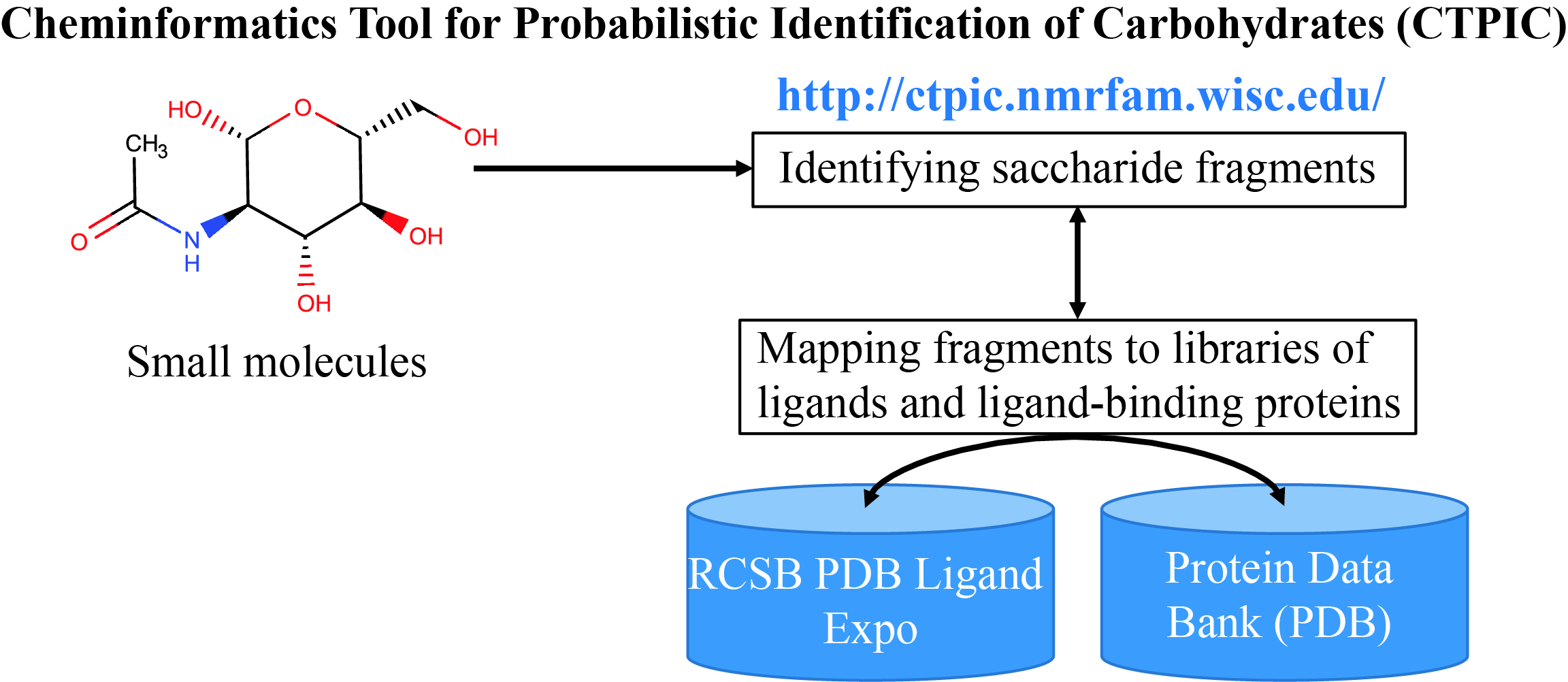
CTPIC
Cheminformatics Tool for Probabilistic Identification of Carbohydrates (CTPIC) analyzes the covalent structure of a chemical compound to yield a probabilistic measure for distinguishing saccharides and saccharide-derivatives from non-saccharide chemical compounds.
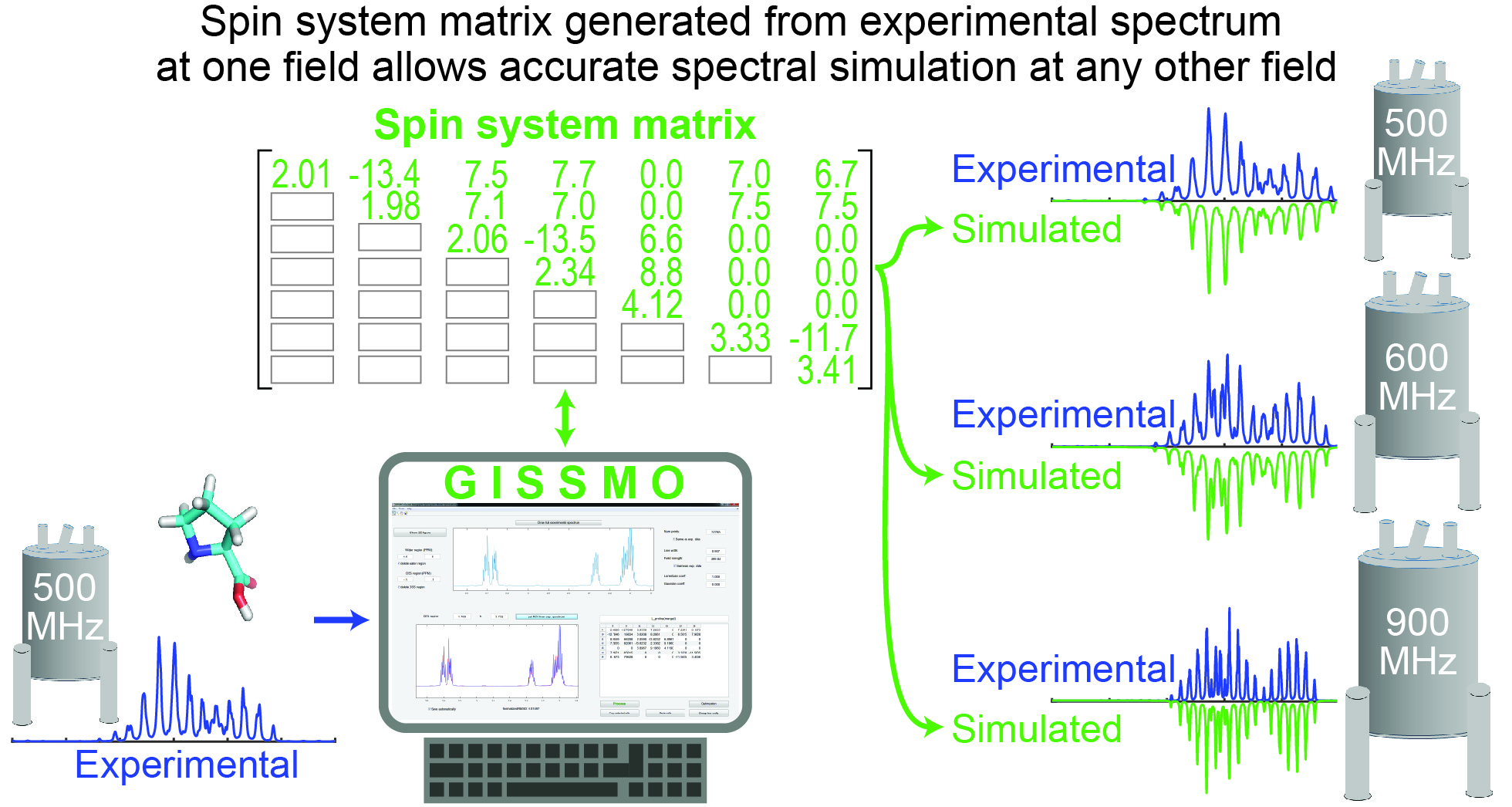
GISSMO
Guided Ideographic Spin System Model Optimization (GISSMO) enables the efficient calculation and refinement of spin system matrices (chemical shift and coupling constants) against experimental 1D-1H NMR spectra of small molecules.
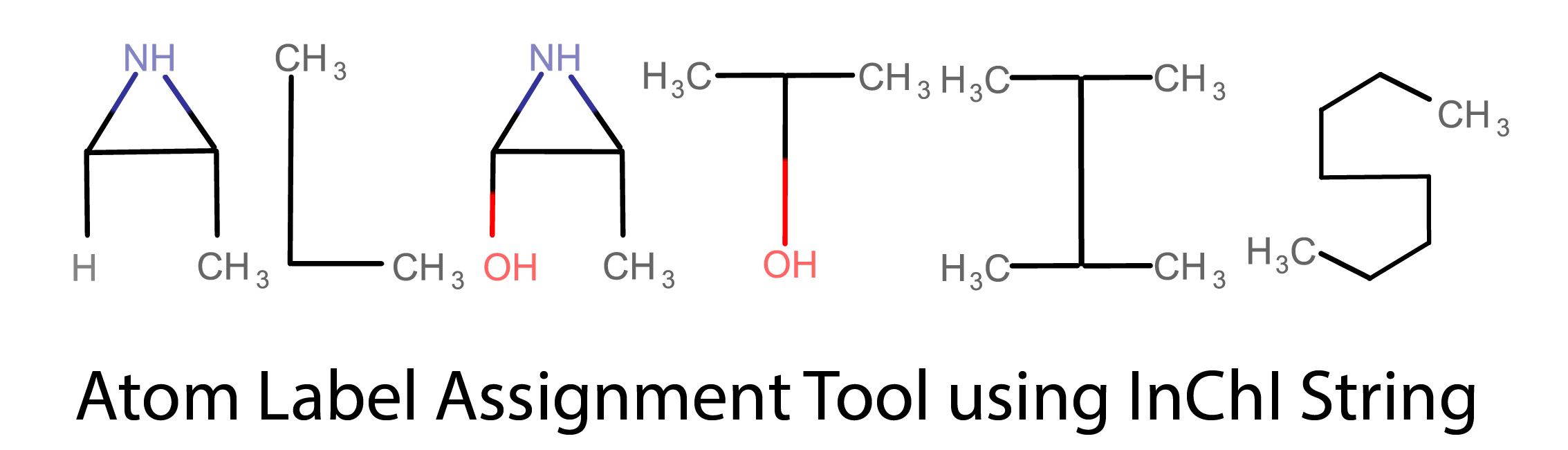
ALATIS
Atom Label Assignment Tool using InChI String (ALATIS) operates fully within the InChI convention to provide unique and reproducible molecule and all atom identifiers.
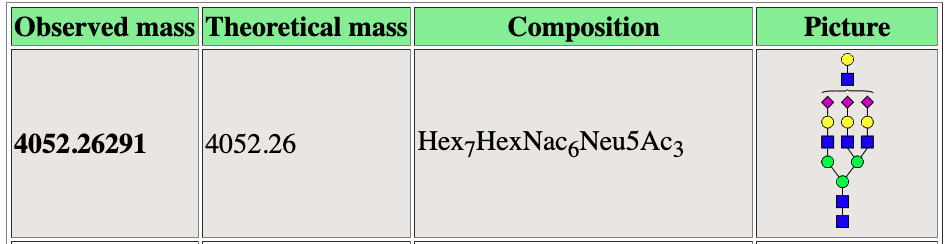
Mass to Comp
Mass2Comp is a webserver for computational assignment of glycan peaks. This is an on going collaboration with Dr. Sylvain Lehoux at The National Center for Functional Glycomics
RUNER
Robust and Unique Nomenclature for Enhanced Reproducibility (RUNER) is a robust and standardized pipeline that enables seamless modifications of atom force field parameters for computational molecular dynamics, energy minimization, and modeling of molecular interactions.
Open positions
Through different collaborations, we have openings for talented research scientists/postdocs/programmers with expertise in one of the areas of data science, machine learning, computational epidemiology, or computational genomics.Send me your CV and your research interest statement.
Funding
Current- Cardiovascular Disease Initiative (CVDi), Broad Institute of MIT and Harvard
- Women's Health Study: Infrastructure support for cohort follow-up supplement U01 CA182913
- Chief’s pilot research funding award, Division of Preventive Medicine, Brigham and Women’s Hospital